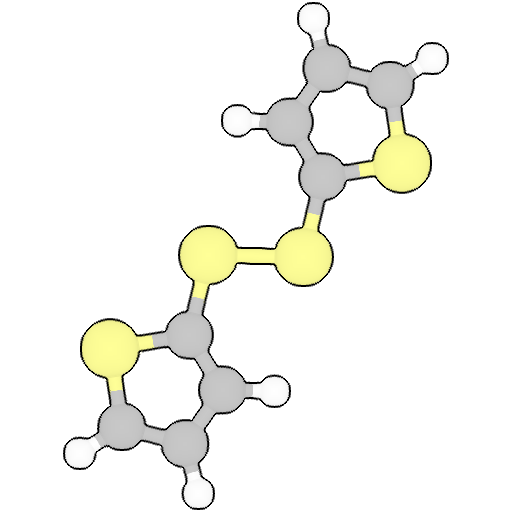
Geometry
Geometry Optimization
tensormol02
Energy: -1901.1222833210488 Ha
1 S -2.898374 -2.793559 0.000871
2 S -0.871812 -2.868608 0.001386
3 S -5.111976 -4.817311 0.100375
4 S 1.343649 -0.844349 0.002376
5 C -4.770975 -6.511928 0.086111
6 C 1.001903 0.850199 0.001669
7 C -3.441897 -6.768432 0.026938
8 C -0.328567 1.106347 0.000550
9 C -2.647042 -5.585849 -0.008708
10 C -1.123894 -0.076437 0.000240
11 C -3.387073 -4.456414 0.023872
12 C -0.382898 -1.205699 0.002466
13 H -5.603965 -7.192078 0.123671
14 H 1.835532 1.530607 0.001794
15 H -3.012139 -7.756120 0.007893
16 H -0.759035 2.093909 -0.000156
17 H -1.572558 -5.578738 -0.056075
18 H -2.199415 -0.083826 -0.002430
of 2
Conformation Search
tensormol02 - 20
Energy: -1900.9677675980072 Ha
1 S -1.133058 0.181288 0.052096
2 S 0.915604 -0.208585 0.138165
3 S -3.448154 -1.726207 0.131387
4 S 2.754802 2.211250 0.054154
5 C -3.163227 -3.429188 0.119639
6 C 3.299088 3.458851 -0.629330
7 C -1.844672 -3.739933 0.059926
8 C 2.038931 3.744110 -0.020549
9 C -1.007571 -2.592027 0.023651
10 C 1.093693 2.574677 0.152289
11 C -1.711135 -1.439614 0.064287
12 C 1.498048 1.248736 0.554046
13 H -4.026047 -4.068116 0.158677
14 H 4.176836 3.935501 -0.923116
15 H -1.454377 -4.743620 0.042545
16 H 1.747046 4.650816 0.505002
17 H 0.064694 -2.631414 -0.037426
18 H 0.199497 2.573475 -0.445443
of 19
symmetry c1
Atomic Coordinates
indices
1 S -2.884425 -2.812627 0.016730
2 S -0.885061 -2.849590 0.014410
3 S -4.955706 -4.781526 0.090724
4 S 1.187256 -0.879995 -0.003240
5 C -4.707380 -6.426344 0.083728
6 C 0.938216 0.764760 0.001364
7 C -3.380022 -6.783545 0.024498
8 C -0.390610 1.121429 0.001063
9 C -2.586916 -5.651915 -0.013479
10 C -1.184127 -0.010487 -0.002353
11 C -3.289269 -4.495817 0.024801
12 C -0.480370 -1.166333 0.003786
13 H -5.595793 -6.986301 0.124675
14 H 1.827323 1.325110 0.003242
15 H -3.097701 -7.832028 0.013180
16 H -0.673618 2.169783 0.002747
17 H -1.498040 -5.679724 -0.066342
18 H -2.274295 0.016862 -0.006691