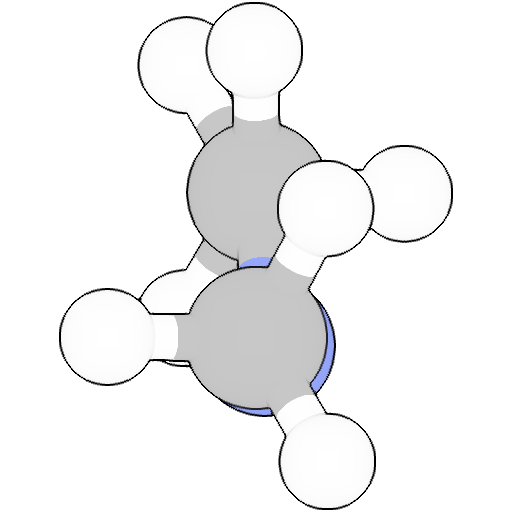
Geometry
Geometry Optimization
tensormol02
Energy: -173.24559964174222 Ha
1 C -0.100555 -0.376186 -0.814162
2 H -0.071049 0.632630 -0.368155
3 N -0.064460 -1.439772 -0.112653
4 C -0.090478 -1.457891 1.336335
5 H -1.117030 -1.477898 1.726661
6 H 0.400727 -2.369565 1.696586
7 H 0.411302 -0.588754 1.786222
8 C -0.081857 -0.453060 -2.316957
9 H 0.951305 -0.463348 -2.686199
10 H -0.551198 -1.389191 -2.643319
11 H -0.585183 0.406141 -2.769647
of 2
Conformation Search
tensormol02 - 100
Energy: -173.2421958642063 Ha
1 C 0.520647 -0.112709 -0.129071
2 H 0.696629 -0.964756 -0.803596
3 N -0.550359 0.012114 0.541719
4 C -1.773560 -0.680205 0.153604
5 H -2.087673 -0.377799 -0.858416
6 H -1.661769 -1.772324 0.119506
7 H -2.594328 -0.439735 0.841760
8 C 1.647673 0.885968 0.005513
9 H 1.267665 1.915280 -0.066969
10 H 2.146873 0.807145 0.977605
11 H 2.388202 0.727020 -0.781655
of 100
symmetry c1
Atomic Coordinates
indices
1 C -0.072615 -0.443319 -0.799892
2 H -0.100079 0.547980 -0.325931
3 N -0.027109 -1.515360 -0.109689
4 C -0.089377 -1.459585 1.357932
5 H -1.126068 -1.453346 1.713377
6 H 0.409901 -2.323303 1.811606
7 H 0.397330 -0.556686 1.743936
8 C -0.072615 -0.443319 -2.339892
9 H 0.948077 -0.452226 -2.738912
10 H -0.590675 -1.322810 -2.738912
11 H -0.575247 0.445080 -2.738912